 ore and more articles analyzing the DNA sequence of SARS-CoV-2 are popping up
all over the Internet. Some dispute the conclusion of that Nature article that
claims it evolved naturally, while others try to dismiss them as “conspiracy
theories.” In fact, most are science blogs like this one (though, I should
point out, not nearly as nice).
ore and more articles analyzing the DNA sequence of SARS-CoV-2 are popping up
all over the Internet. Some dispute the conclusion of that Nature article that
claims it evolved naturally, while others try to dismiss them as “conspiracy
theories.” In fact, most are science blogs like this one (though, I should
point out, not nearly as nice).
For the most part, these are not written by professional virologists, who would face serious career repercussions for making speculative reports, but individuals who are highly knowledgeable about molecular biology. Given what's happening, most naturally wish to remain anonymous.
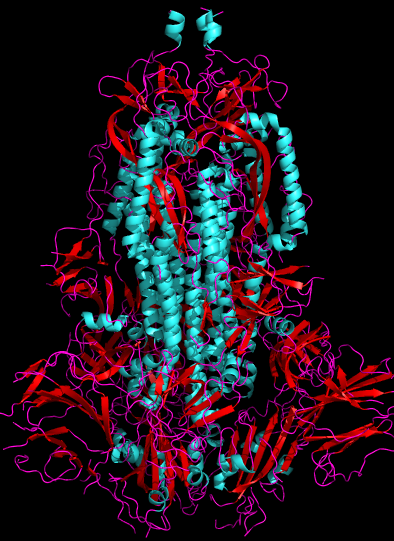
Fig. 1. 2.8Å cryo-electron microscopy 3D structure of SARS-CoV-2 spike glycoprotein[1] (6VXX), side view. Human ACE2-binding SB domains are at the bottom. Viral membrane would be at the top. Cyan = alpha-helix, Red = beta-sheet, Purple = random chain
When legitimate questions by laymen are misrepresented and “debunked” we end up with confusion. It doesn't help when their conclusions are echoed by scientifically naive political commentators on disreputable sites who don't fully understand the science.
But they help to spur interest and excitement about molecular biology. This is something to be encouraged. Finding the origin of this virus is not only legitimate, it is essential. These people could get Ph.D.s, if they don't have them already, and then start competing with me for grants. Which, now that I think of it, is bad!
Viruses are far and away the most deadly pathogens on the planet. It's been estimated that every two days, viruses kill half of all the bacteria on the planet.[7] That's equal to one fourth of all living things killed by viruses every 48 hours. Hepatitis B virus kills a million humans a year, as does measles, in part to its immunosuppressive effect. Human papilloma virus (HPV), the most common sexually transmitted infection, is the second biggest cause of cancer in women, causing 500,000 cases of cervical cancer every year, half of which are fatal. Many other viruses kill comparable numbers.
In this article I'll take a look at what these bloggers are saying and compare it to what is known about SARS-CoV-2 and in particular about S, the spike glycoprotein.
Furin cleavage site
The spike glycoprotein has a trimeric structure[1] and covers the surface of SARS-CoV-2 like spikes on a mace. It contains two subunits: S1, which contains the receptor binding domain, and S2, which is responsible for fusion of the viral and cellular membranes. The point where the subunits are joined is called the S1/S2 site. Proteolysis of this site is essential for infection.
Some researchers proposed that an additional site called S2′ within the S2 protein (at R797 in SARS-CoV) is necessary to expose the viral fusion peptide, which is a relatively apolar region of 15–25 amino acids that curves the target membrane to drive the membrane fusion reaction.[2] Hemagglutinin serves this important function in influenza virus.
The S1 protein has a V-shape that can be “open” or partially “closed” and spontaneously changes from one conformation to the other. In the open conformation, the hACE2 binding motifs (called SB) are exposed. The virus binds to the cell and undergoes conformational changes leading to cleavage of the S2′ site, which allows it to fuse to the membrane and enter the cell. This is different from coronaviruses that cause colds, which remain in the closed configuration, and may account for the greater pathogenicity of SARS-type viruses.
The S trimer also has 66 N-linked glycans on the surface (22 per protomer), mainly N-acetyl-D-glucosamine or NAG, which participate in S folding and affect how it interacts with proteases, which is to say they help determine whether it can infect a human cell. They also make generating antibodies against it harder.
The S1/S2 site in SARS-CoV-2 is highly unusual in that it contains two protease sites: an RS (arginine-serine) motif for TMPRSS2 (and possibly cathepsin) and a RRAR motif for furin. Furin is an abundant protease found in human cells but not in viruses. Influenza viruses have a furin cleavage site, and it's one reason why influenza is so contagious. Other SARS-type viruses don't have it, and its appearance is taken by some skeptics as possible evidence of protein engineering.
The extent to which the furin site in SARS-CoV-2 enhances pathogenicity is not yet clear, but it's believed to expand the tropism of the virus (which means it can infect more different types of cells). Walls et al.[1] deleted the four-amino-acid furin insert (in a pseudovirus) by mutating TNSPRRAR to TILR and found that some types of cells were infected more easily and some less easily. They conclude:
We speculate that the almost ubiquitous expression of furin-like proteases could participate in expanding SARS-CoV-2 cell and tissue tropism, relative to SARS-CoV, as well as increasing its transmissibility and/or altering its pathogenicity.
What this means is that SARS-CoV-2 could potentially get into almost any cell that has an ACE2 protein. This might explain some of the non-respiratory symptoms that have been seen in COVID-19 patients.
Needless to say, nailing down this point will be a critical task for drug developers.
Here's a cartoon rendering of the SARS-CoV-2 spike glycoprotein obtained by electron microscopy. Alpha-helices are in blue, beta-sheets are rendered as flat arrows, and random chains are in purple.
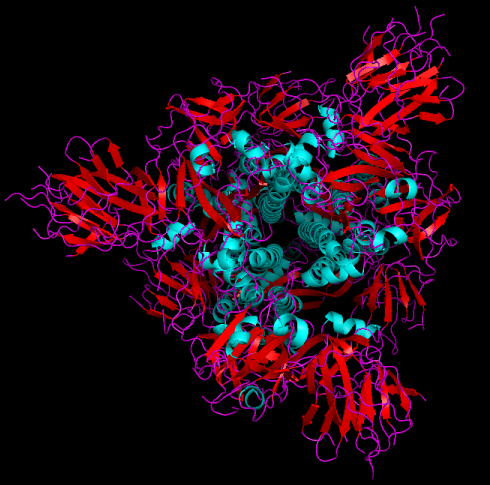
Fig. 2. 3.46Å cryo-electron microscopy 3D structure of SARS-CoV-2 spike glycoprotein (6VSB)[3] rendered in pymol (top view showing three-fold symmetry).
The part we're interested in is the S1/S2 cleavage point at the junction point between its two halves (S1 and S2), that can be cut by TMPRSS2. This is shown below. Amino acids 677–688, which contain the S1/S2 cleavage sites, are missing from the EM structure, apparently because the authors abrogated the site before doing the cryo-EM. Many other regions are also unresolved due to conformational heterogeneity. So the protein in the database is not good enough to run a docking model. But it does tell us that the cleavage site is on the surface of the protein and accessible to solvent. If it were buried somewhere inside the protein, it would be non-functional and just a curiosity.
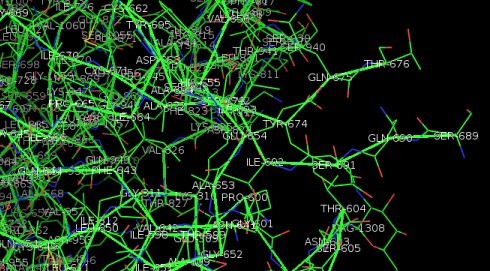
Fig. 3. Stick diagram derived from 2.8Å cryo-electron microscopy 3D structure of SARS-CoV-2 spike glycoprotein[1] (6VXX), close-up and annotated to show cleavage site exposed on surface
Shi's Nature paper[4] reported the peptide sequence of S1, but it ends at residue 675—almost exactly where the furin site begins. Here's the region of interest.
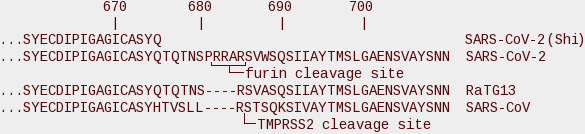
The top sequence is what was reported in Shi's paper. The second line shows the complete sequence around the cleavage site, and the third line shows RaTG13, the putative ancestor of SARS-CoV-2. TMPRSS2 cleaves at the RS motif at 685/686, cutting the spike glycoprotein roughly in half and releasing the free S1 and S2 subunits. Furin binds at an RxxR site (where R is arginine and x is any amino acid). If a second R is present (RRxR or RxRR) the cleavage efficiency is greatly enhanced. SARS-CoV-2 has such a site (RRAR) thanks to the PRRA insert appearing immediately before the RS cleavage site. As shown above, this site is solvated and it's in an ideal location for furin to cleave.
Furin cleaves after the arginine (i.e. R↓S)[9]. Furin cleaves many other viral membrane fusion proteins and pro-toxins, including anthrax, Shiga, diphtheria, and botulinum toxins and influenza A H5N1, Marburg, HIV, and Ebola viruses. Furin cleavage of influenza hemagglutinin drastically increases its pathogenicity.[10]
The closest relative and putative ancestor, RaTG13, does not have a furin-binding site, nor does SARS-CoV or MERS. The furin insertion—four amino acids—can't happen by point mutation, so it's extremely unlikely that this furin binding site could have evolved from SARS.
Understanding the furin site would be vital to someone trying to repurpose protease inhibitors as drugs. The discovery that a ubiquitous protease like furin cleaves the spike protein means that all those papers talking about how pathogenicity depends on TMPRSS2 and developing inhibitors for it[5] may be futile. It's one reason why drug development in cell culture so often fails.
The exact sequence seems suspicious to some bloggers because the virus would not only have to undergo recombination, but also a mutation. One writes:
Although similar furin-cleavage sites have been observed in other coronaviruses, none of them contains the same exact sequence. Therefore, the chance that the furin-cleavage site in the Wuhan coronavirus was obtained through recombination with another furin-cleavage-site-containing coronavirus is very low.
Assuming it wasn't cloned into the DNA (which is fairly easy to do these days), the only way for it to get there is if two different viruses happen to infect the same cell at the same time. Since RaTG13 has never been documented in humans, this means it either happened in a bat that contracted RaTG13 and human-type influenza simultaneously, or in cell culture. Both are possible. It's even possible that SARS-CoV-2 was accidentally created in a lab worker without anyone realizing.
I could go on, but Yuri Deigin's article has a lot more detail about how researchers have taken risks to help our understanding of these viruses, including a reference to a preprint that suggests that Shi isolated RaTG13 in 2013 from the horseshoe bat Rhinolophus affinis and named it RaBtCoV/4991 (GenBank KP876546) in 2016.[6] Although only a short piece of it was sequenced, it's identical to RaTG13. This dumps cold water on the theory that Shi fabricated the RaTG13 sequence. Instead it suggests that there could be many other strains similar to RaTG13 and SARS-CoV-2 out there. If so, that's something we need to know.
Questioning the origin of RatG13
A second blogger hypothesizes that RatG13 never existed, but was invented by Shi's lab to cover their tracks after SARS-CoV-2 escaped.
Needless to say this biologist, whose writing style marks him as a native Chinese speaker, remains strictly anonymous and refuses all attempts to get him to reveal his email address or institutional affiliation. If he did, he would likely “disappear.” He writes:
So the RaTG13 virus, if indeed exists, should be able to infect humans. I said in my article that Zhengli Shi needed to take one little peek at the sequence of RaTG13 and realize at once that this virus has the potential to infect humans. There is no reason that she should let this thing sit for 7 years and only decided to publish its sequence once the outbreaks took place.
The RaTG13 S1 gene used in this Nature article was synthesized, not obtained from Zhengli Shi (they have been collaborators in the past), a proof of my other claim --- Shi does not have a physical copy of the RaTG13 virus.
On this point, the WIV recently stated that they only have samples of three viruses at the moment and none of them are closely related to SARS-CoV-2. But it's wild speculation to say it was invented. It's not necessary to have a sample of a live virus these days. Many viruses have been completely sequenced and characterized without ever growing a sample in culture. Cultivating live viruses is orders of magnitude harder than sequencing some DNA found in bat excreta.
What's the truth?
Of course, bloggers can only speculate, but especially given the existence of RaBtCoV/4991, it seems extremely unlikely that the RaTG13 sequence is fake. As most of the bloggers (including the two cited above) repeatedly point out, there's no way to tell from the DNA or peptide sequence where something came from—nobody leaves EcoR1 restriction sites flanking their insert behind anymore like we did in the 1980s. Blunt-end cloning and PCR cloning are commonplace now.
As mentioned above, it has also not been established how much furin cleavage contributes to pathogenicity. This is something that we need to know before we can design effective protease inhibitors. Potent inhibitors of furin exist, but haven't been tested on COVID-19, and their biological effects would be hard to evaluate due to the large numbers of substrates. It would be nice to have an X-ray structure that includes the cleavage site.
An investigation into exactly what went wrong in Wuhan is necessary and inevitable. So far the PRC government is being uncooperative: when Australia last week called for an investigation, and 100 other countries signed up, the PRC threw up a massive tariff on Australian barley in retaliation. More recently they've insisted that the WHO, which has uncritically repeated false statements provided by Beijing in the past, be in charge.
This is China's opportunity to prove to the world that they can behave like a modern open society. Hopefully they will take it.
Update May 21 2020 A BiorXiv preprint[8] from the University of Pittsburgh presents evidence that the PRRA insert unique to SARS-CoV-2 has close structural similarity to the SEB superantigen, which is a highly immunogenic sequence in bacteria. They suggest that this insert could be responsible for the MIS-C (Kawasaki-like) symptoms in children and the cytokine storm in adults and say antibodies against the S1/S2 domain might be more effective than antibodies against the receptor-binding domain, which most people are making.
Here are some Genbank accession numbers:
DQ514532.1 (locus ABF68959.1) SARS CoV CS24 spike glycoprotein [Wuhan].
ABD72996.1 (locus ABD72996) Hong Kong SARS-related CoV spike glycoprotein
YP_009724390.1 (locus YP_009724390) (SARS-CoV2 surface glycoprotein, Shanghai
7BV2_A (7BV2_A) SARS-CoV2 pdb: molecule 7BV2, chain 65, EM structure
MN996532.1 (locus QHR63300.2) Spike glycoprotein [Bat CoV RaTG13], Wuhan
KP876546 the mysterious RaBtCoV/4991
Here are some RDB numbers:
6VXX 2.8Å EM 3D structure of SARS-CoV-2 Spike Glycoprotein.
6VSB 3.46Å EM 3D structure of SARS-CoV-2
Spike Glycoprotein with a single receptor-binding domain up
Here are some links. Grab copies before they get censored.
Yuri
Deigin's analysis on medium.com
Jean-Claude Perez (2020).
Wuhan covid-19 synthetic origins and evolution. International Journal of Research -
Granthaalayah, 8(2), 285–324 (Zenodo.org preprint)
Anonymous
virologist at nerdhaspower.weebly.com
1. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181(2), 281–292. doi: 10.1016/j.cell.2020.02.058. PMID: 32155444
2. Madu IG, Roth SL, Belouzard S, Whittaker GR (2009). Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 83(15), 7411–7421. doi: 10.1128/JVI.00079-09. PMID: 19439480
3. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C, Abiona O, Graham BS, McLellan JS (2020). Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up. Science 367, 1260–1263.
4. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798), 270–273. doi: 10.1038/s41586-020-2012-7. https://www.ncbi.nlm.nih.gov/pubmed/32015507 PMID: 32015507 PMCID: PMC7095418
5. Hoffmann M, Kleine-Weber H, Schroeder S, Kröger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181(2), 271–280. doi: 10.1016/j.cell.2020.02.052. PMID: 32142651
6. Ge X, Wang N, Zhang W, Hu B, Li B, Zhang Y, Zhou J, Luo C, Yang X, Wu L, Wang B, Zhang Y, Li Z, Shi Z (2016). Coexistence of Multiple Coronaviruses in Several Bat Colonies in an Abandoned Mineshaft Virol Sin 31(1), 31–40. doi: 10.1007/s12250-016-3713-9. PMID: 26920708 PMCID: PMC7090819 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7090819/pdf/12250_2016_Article_3713.pdf
7. Norkin LC (2010). Virology: Molecular Biology and Pathogenesis p.39
8. Cheng, M. H., Zhang, S., Porritt, R. A., Arditi, M., Bahar, I. (2020) An insertion unique to SARS-CoV-2 exhibits superantigenic character strengthened by recent mutations. doi 10.1101/2020.05.21.109272 Biorxiv preprint, not yet peer reviewed, Posted: 2020-05-21 https://www.biorxiv.org/content/10.1101/2020.05.21.109272v1
9. Remacle AG, Shiryaev SA, Oh ES, Cieplak P, Srinivasan A, Wei G, Liddington RC, Ratnikov BI, Parent A, Desjardins R, Day R, Smith JW, Lebl M, Strongin AY (2008). Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 283(30), 20897–20906. doi: 10.1074/jbc.M803762200. PMID: 18505722
10. Braun E, Sauter D (2019). Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunology. 8(8), e1073. doi: 10.1002/cti2.1073 PMCID: PMC6682551 PMID: 31406574
Hat tip to a reader who alerted me to the Nerdhaspower article.
may 21 2020, 7:33 am. minor edits 2:43 pm. Updated Friday, May 22, 2020, 6:45 pm. last edited jun 06 2020, 2:30 pm
